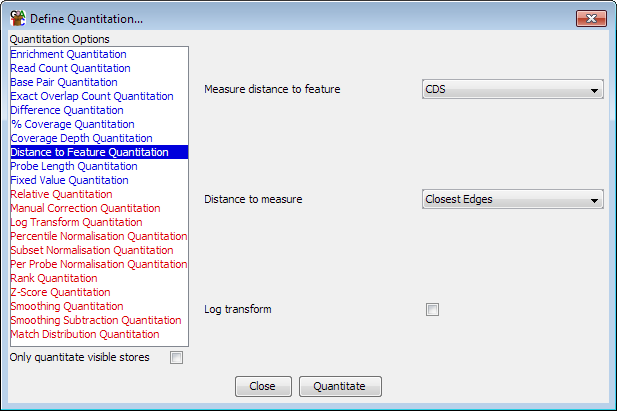
This quantitation methods simply quantitates each probe with the distance to the nearest member of a class of selected features. Since the quantitation doesn't depend on the data in each data store every data store will get the same quantitated values.

You can choose whereabouts in the probe you want to start your measurement and where in the feature it should end. The options are either to measure from the centre of the probe to the centre of the feature, or to measure the distances between their closest edges. All distances are absolute (no negative distances allowed). You can also opt to log2 transform your distances if the distances involved are going to be large.
Any probe on a chromosome which doesn't contain an instance of the feature being searched against will be assigned the length of the chromosome.